-
NGS is a cost-effective technology for the identification of Genetic variants affecting a gene or region of the genome.
NGS – Next-generation sequencing
Technology designed to detect DNA sequence variations in a gene or genomic region


NGS is a cost-effective technology for the identification of Genetic variants affecting a gene or region of the genome.
Test results are interpreted based on the recommendations and guidelines of the American College of Medical Genetics and Genomics (ACMG) and the European Society of Human Genetics (ESHG) as described below.
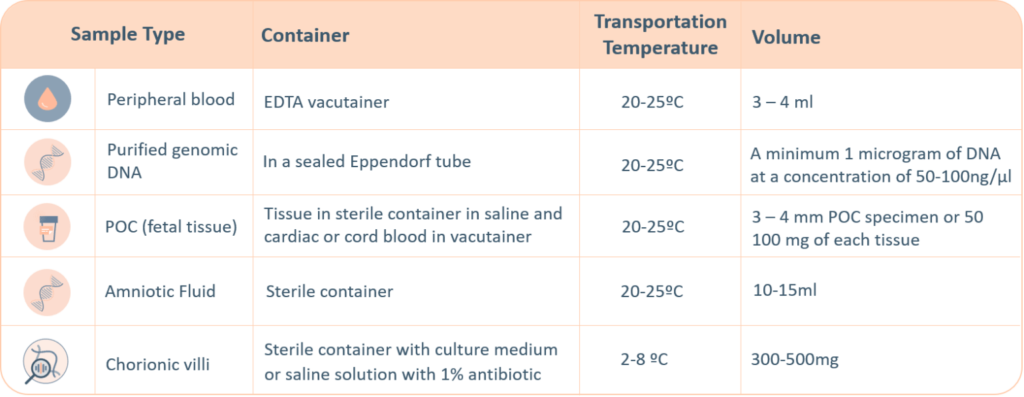
A thorough labelling of the tube with unique identifying information is required, incorrect labelling can lead to rejection of the sample. The minimum required information to identify and accept a sample is: Patient’s full name, Date of birth, Gender and Medical Record Number.
The ‘informed consent’ and the ‘test requisition from’ must be properly filled-in and signed by the patient and doctor.